We proudly announce the release of RQGIS! RQGIS establishes an interface between R and QGIS, i.e. it allows the user to access the more than 1000 QGIS geoalgorithms from within R. To install it, run:
install.packages("RQGIS")Naturally, you need to install other software (such as QGIS) to run RQGIS properly. Therefore, we wrote a package vignette, which guides you through the installation process of QGIS, GRASS and SAGA on various platforms. To access it, run:
vignette("install_guide", package = "RQGIS")How to use RQGIS
To introduce RQGIS, we will demonstrate how to calculate the SAGA topographic wetness index. The first step is the creation of a QGIS environment. This is a list containing the paths RQGIS needs to access QGIS. Luckily for us, set_env does this for us. We merely need to specify the root path to the QGIS installation. This is most likely something like C:/OSGeo4W~1 on Windows, /usr on Linux and /usr/local/Cellar on a Mac. If we do not specify a path to the QGIS root folder, set_env tries to figure it out. This, however, might be time-consuming depending on the size of the drives to search.
library("RQGIS")
env <- set_env()Secondly, we need to find out the name of the function in QGIS which calculates the wetness index:
find_algorithms("wetness index", name_only = TRUE, qgis_env = env)## [1] "taudem:topographicwetnessindex" "saga:sagawetnessindex"
## [3] "saga:topographicwetnessindextwi" ""There are three algorithms containing the words wetness and index in their short description. Here, we choose saga:sagawetnessindex. To retrieve the corresponding function arguments, we use get_args_man. By setting option to TRUE, we indicate that we would like to use the default values, if available:
args <- get_args_man(alg = "saga:sagawetnessindex",
options = TRUE,
qgis_env = env)
# print the retrieved list
args## $DEM
## [1] "None"
##
## $SUCTION
## [1] "10.0"
##
## $AREA_TYPE
## [1] "0"
##
## $SLOPE_TYPE
## [1] "0"
##
## $SLOPE_MIN
## [1] "0.0"
##
## $SLOPE_OFF
## [1] "0.1"
##
## $SLOPE_WEIGHT
## [1] "1.0"
##
## $AREA
## [1] "None"
##
## $SLOPE
## [1] "None"
##
## $AREA_MOD
## [1] "None"
##
## $TWI
## [1] "None"Of course, we need to specify certain function arguments such as the input (DEM) and output (TWI) arguments. Please note that RQGIS accepts as input argument either the path to a spatial object or a spatial object residing in R. Here, we will use a digital elevation model, which comes with the RQGIS package:
# load data into R
data("dem", package = "RQGIS")
# define input
args$DEM <- dem
# specify output path
args$TWI <- "twi.sdat"Finally, we can access QGIS from within R by supplying run_qgis with the specified arguments as a list. Specifying also the load_output-argument directly loads the QGIS output into R.
twi <- run_qgis(alg = "saga:sagawetnessindex",
params = args,
load_output = args$TWI,
qgis_env = env)
# visualize the result
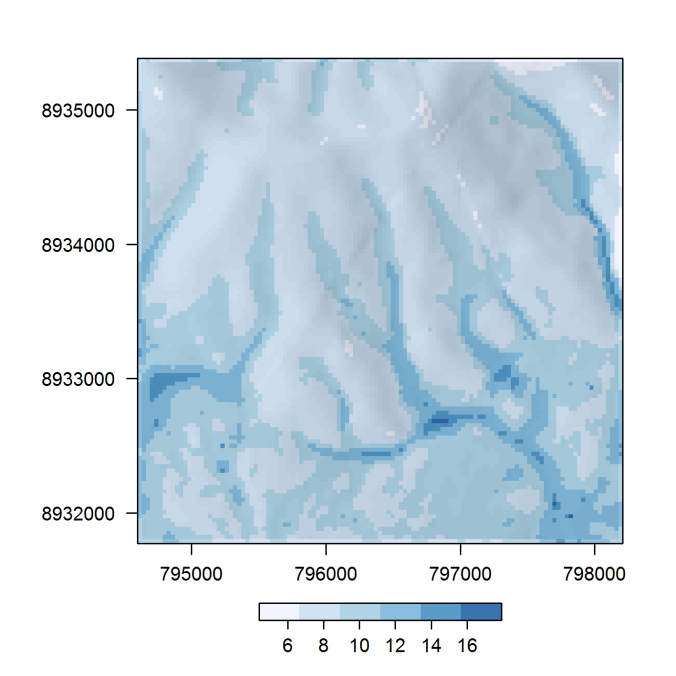
library("raster")
hs <- hillShade(terrain(dem), terrain(dem, "aspect"), 40, 270)
pal <- RColorBrewer::brewer.pal(6, "Blues")
spplot(twi, col.regions = pal, alpha.regions = 0.8,
scales = list(tck = c(1, 0)),
colorkey = list(space = "bottom",
width = 1, height = 0.5,
axis.line = list(col = "black")),
cuts = length(pal) - 1) +
latticeExtra::as.layer(spplot(hs, col.regions = gray(0:100 / 100)),
under = TRUE)
For more information on RQGIS, please refer to https://github.com/jannes-m/RQGIS.
Pingback: RQGIS 0.1.0 release | A bunch of data
i1m very happy with this release. Thank you guys!!!!
I’ve been intersecting heavy geometries and with each method it took TOO LONG, so I’m trully happy with this. If i can help you with something just let me know
LikeLike
Pingback: RQGIS 0.1.0 release – Mubashir Qasim
Having to let RQGIS search for the OSGeo4W is pretty tedious.
Can we just set our own environment variable on Windows, like:
QGIS_HOME=c:\bin\QGIS2.16.1
-or-
OSGeo4W_HOME=c:\bin\QGIS2.16.1
??
Thanks for your work on this.
LikeLike
Yes, you can specify the root path to the QGIS-installation yourself, i.e. in your case probably:
my_env <- set_env("C:/bin/QGIS2.16.1")Subsequently, you have to provide other functions with the my_env-object, e.g.,
find_algorithms(search_term = "wetness", qgis_env = my_env)Please refer also to the documentation (?set_env) and the example found on https://github.com/jannes-m/RQGIS.
LikeLike
I have another question, please.
I setwd() to a directory where I have spatial code and files for R (a projects dir).
If I execute run_qgis(….) with the arguments, and twi.asc does not exist, I get:
Error in .rasterFromASCIIFile(x, …) :
C:\/twi.asc does not exist
If I create the file in the , leaving it empty, I get:
Error in matrix(unlist(ini, use.names = FALSE), ncol = 2, byrow = TRUE) :
‘data’ must be of a vector type, was ‘NULL’.
How does twi.asc get populated, by running run_qgis(….) ?
LikeLike
It seems you are using QGIS 2.16.1. This release has a minor bug. I explain how to fix it on https://github.com/jannes-m/RQGIS in section “QGIS 2.16 modifications”. To avoid the manual adjustment we recommend to use the QGIS long term release (2.14). Using the OsGeo4W-installer, you can install both the latest (2.16.1) and the long term release (2.14). In this case RQGIS will automatically use the LTR. For a detailed installation guide please refer to our package vignette (
vignette("install_guide", package = "RQGIS")).LikeLike
Thank you very much, I want only to notice that another likely location of Qgis in Mac is
/Applications/QGIS.app
as env <- set_env() was able to figure in my machine.
Development version from dakota carto
env <- set_env("/Applications/QGIS_2.15-dev.app")
LikeLike
Hi, thanks for creating the RQGIS package I have been waiting it for so long! I’m trying to get into it. While I was able to extract centroids from one of my shapefiles (following the code from the README file in Gitub), I’m having some problems to merge a couple of layers. I think I’m not defining correctly the ParameterMultipleInput. I tried several ways: defining a folder were the shapefiles to be merge are; reading them into R and calling them directly but nothing is working. Can you give any hint about it?
Thanks in advance!
I’m using QGIS 2.14.2 and R 3.3.1
Here the R code in using:
### EJERCICIO 2: MERGE TWO SPDF
library(“RQGIS”)
library(rgdal)
setwd(“C:\\Users\\jgaleano\\Desktop\\CLASES2014\\CLASE4\\BASES”)
env <- set_env()
bcn <- readOGR(".", "Barcelona")
gir <- readOGR(".", "Girona")
find_algorithms("merge", name_only = TRUE, qgis_env = env) # buscar el algorítmo en QGIS
get_usage(alg = "qgis:mergevectorlayers",
qgis_env = env,
intern = TRUE) # CONOCER LOS COMPONENTES DEL ALGORITNO
params <- get_args_man(alg = "qgis:mergevectorlayers",
qgis_env = env) # CREAR UNA LISTA CON LOS COMPONENTES DEL ALGORITMO
params
params$LAYERS <- c(bcn,gir) # DEFINIR LOS PARAMETROS DEL ALGORITMO
params$OUTPUT <- file.path("C:\\Users\\jgaleano\\Desktop\\TERE", "BARCELONA_GIRONA_MERGE.shp")
out <- run_qgis(alg = "qgis:mergevectorlayers",
params = params,
load_output = params$OUTPUT_LAYER,
qgis_env = env)
plot(out)
LikeLike
Hola Juan,
in your case it would be better not to load your shapefiles directly into R, instead specify the LAYERS paramter as follows (I simply assume your shapefiles can be found on your D: drive, please make sure to indicate the full path!!!):
qgis_env <- set_env("C:/OSGeo4W64/")
params <- get_args_man("qgis:mergevectorlayers", qgis_env = qgis_env)
params$LAYERS <- "D:/Barcelona.shp;D:/Girona.shp"
params$OUTPUT <- "D:/merge.shp"
out <- run_qgis(alg = "qgis:mergevectorlayers",check_params = FALSE,
params = params,
# only indicate the load_output argument if you want to load the QGIS
# output directly into R
load_output = params$OUTPUT,
qgis_env = qgis_env)
Just two more remarks – first of all, it would be of great help if you provided your problem with a reproducible example. And secondly, post your problem on GIS exchange (http://gis.stackexchange.com/questions) or submit it to the r-sig-geo list (http://r-sig-geo.2731867.n2.nabble.com/) since this will ensure that more people could help you. Additionally, if another person has a similar problem to yours in the future, it will be easier for him/her to find the answer there.
Cheers,
Jannes
LikeLike
Pingback: Esta es una estupenda noticia – R+GIS